13.13 Energy Decomposition Analysis based on SAPT/cDFT
Many schemes for decomposing quantum chemical calculations of intermolecular interaction energies into physically meaningful components can be found in the literature, but the definition of the charge-transfer (CT) contribution has proven particularly vexing to define in a satisfactory way and typically depends strongly on the choice of basis set,[Ronca et al.(2014)Ronca, Belpassi, and Tarantelli, Řezáč and de la Lande(2015), Lao and Herbert(2016)] because as virtual orbitals on monomer 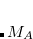 start to extend significantly over monomer
start to extend significantly over monomer  as the basis set approaches completeness, the distinction between polarization (excitations localized on
as the basis set approaches completeness, the distinction between polarization (excitations localized on 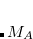 , introduced by the perturbing influence of
, introduced by the perturbing influence of  ) and CT (excitations from
) and CT (excitations from 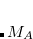 to
to 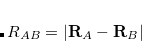 ) becomes blurred.[Lao and Herbert(2016)] This ambiguity renders orbital-dependent definitions of CT highly dependent on the choice of atomic orbital basis set. On the other hand, constrained density functional theory (cDFT, Section 5.13), where a CT-free reference state can be defined based on “promolecule” densities, affords a definition of CT that is scarcely dependent on the basis set and is in accord with chemical intuition in simple cases.[Lao and Herbert(2016)]
) becomes blurred.[Lao and Herbert(2016)] This ambiguity renders orbital-dependent definitions of CT highly dependent on the choice of atomic orbital basis set. On the other hand, constrained density functional theory (cDFT, Section 5.13), where a CT-free reference state can be defined based on “promolecule” densities, affords a definition of CT that is scarcely dependent on the basis set and is in accord with chemical intuition in simple cases.[Lao and Herbert(2016)]
For intermolecular interactions, the cDFT definition of CT can be combined with a definition of the remaining components of the interaction energy (electrostatics, induction, Pauli repulsion, and van der Waals interactions) based on symmetry-adapted perturbation theory (SAPT, Section 13.11). In traditional SAPT, the CT interaction energy resides within the induction energy (also known as the polarization energy), which is therefore itself highly dependent upon the basis set. However, using cDFT to define the CT component and subtracting this out of the SAPT induction energy, both the CT and the remaining induction energies are largely independent of basis set.[Lao and Herbert(2016)] SAPT/cDFT therefore provides a stable and physically-motivated energy decomposition, which can be invoked by setting the $rem variable SAPT_CDFT_EDA = TRUE in a SAPT calculation. A $cdft section must be set to specify the monomer charges and spins for the cDFT calculation.
While the cDFT definition of CT exhibits only a very mild basis-set dependence, its quantitative details do depend upon how the charge constraints in cDFT are defined relative to fragment populations (Section 5.13). For SAPT/cDFT, both atomic Becke[Becke(1988a)] and fragment-based Hirshfeld[Řezáč and de la Lande(2015)] (FBH) charge partitioning methods are available. The former involves construction of atomic cell functions that amount to smoothed Voronoi polyhedra centered about each atom. A switching function defines the atomic cell of atom  , and falls rapidly from
, and falls rapidly from 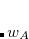 near the nucleus for atom
near the nucleus for atom  , to
, to  near any other nucleus. Becke[Becke(1988a)] defined atomic cell functions
near any other nucleus. Becke[Becke(1988a)] defined atomic cell functions  that are products of switching functions and that can be used to define the cDFT integration weight for monomer
that are products of switching functions and that can be used to define the cDFT integration weight for monomer 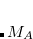 by summing over atoms
by summing over atoms  :
:
 |
(13.39) |
The sum in the denominator runs over all atoms in both monomers, 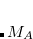 and
and 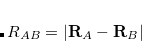 . Becke populations, however, are rooted in a somewhat arbitrarily-defined topology, based in part on assumed atomic radii, whereas FBH partitioning derives physical significance from isolated monomer densities
. Becke populations, however, are rooted in a somewhat arbitrarily-defined topology, based in part on assumed atomic radii, whereas FBH partitioning derives physical significance from isolated monomer densities  and
and  . The cDFT weight function for monomer
. The cDFT weight function for monomer 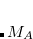 is[Řezáč and de la Lande(2015)]
is[Řezáč and de la Lande(2015)]
 |
(13.40) |
which is the same “stockholder” scheme used to define atomic Hirshfeld populations (Section 11.2.1), but applied here to the entire monomer. In the language of cDFT, the denominator in this expression would be called the promolecule density for the dimer  . In order to set a molecular fragment constraint, simply retain the existing syntax in the $cdft input section (as described in Section 5.13) and specify all atoms within a given molecular fragment.
. In order to set a molecular fragment constraint, simply retain the existing syntax in the $cdft input section (as described in Section 5.13) and specify all atoms within a given molecular fragment.
To perform SAPT/cDFT energy decomposition analysis, the user must request a normal SAPT calculation and in addition set SAPT_CDFT_EDA = TRUE. Users of this method are asked to cite Ref. Lao:2016b.
SAPT_CDFT_EDA
Request a SAPT/cDFT energy decomposition analysis
TYPE:
BOOLEAN
DEFAULT:
FALSE
OPTIONS:
TRUE
Run a SAPT/cDFT calculation.
FALSE
Do not run SAPT/cDFT.
RECOMMENDATION:
None
CDFT_POP
Sets the charge partitioning scheme for cDFT in SAPT/cDFT
TYPE:
STRING
DEFAULT:
FBH
OPTIONS:
FBH
Fragment-Based Hirshfeld partitioning
BECKE
Atomic Becke partitioning
RECOMMENDATION:
None
Example 13.334 Energy decomposition analysis for the water dimer using AO-SAPT+aiD3/cDFT.
$comment
a $cdft input section must be set to specify the monomer
charges and spins for the cDFT calculation.
$end
$rem
SYM_IGNORE true
EXCHANGE gen
BASIS aug-cc-pvdz
XPOL true ! must be set to true for sapt jobs too
XPOL_MPOL_ORDER gas ! gas or charges
XPOL_OMEGA true
XPOL_PRINT 3
SAPT_PRINT 3
SAPT true
SAPT_AO true
SAPT_ORDER 2 ! can be set to 1, ELST or RSPT
SAPT_BASIS dimer ! monomer, dimer (if only 2 monomers), or projected
SAPT_DISP_CORR true
SAPT_DISP_VERSION 3
LRC_DFT true
SAPT_CDFT_EDA true
CDFT_POP fbh ! Fragment-Based Hirshfeld (FBH) charge partitioning
$end
$xc_functional
x wPBE 1.0
c PBE 1.0
$end
$lrc_omega
500
500
$end
$cdft
0
1 1 3
0
1 1 3 s
$end
$molecule
0 1
--
0 1
O -0.702196054 -0.056060256 0.009942262
H -1.022193224 0.846775782 -0.011488714
H 0.257521062 0.042121496 0.005218999
--
0 1
O 2.220871067 0.026716792 0.000620476
H 2.597492682 -0.411663274 0.766744858
H 2.593135384 -0.449496183 -0.744782026
$end